Home page
Taxonomy
Genomics
Transcriptomics
Proteomics
Metabolomics
Phenomics
Breeding
Literature
Software copyright
1. Top navigation bar
The top navigation bar contains the basic functionality of WLP, including links to all omics or tool modules.

2. About
This section includes About WLP, Develpers, News, Vistors, Fundings, Tutorial, Key Refs, Cite us, etc. Users can click different buttons to view the relevant information of the database development.
(1) About WLP: The purpose of WLP and related omics are introduced to users.
 (2) Developer: List of developers, if the user has a question to communicate with the developer, you can move the mouse to a developer's avatar to get its mailbox.
(2) Developer: List of developers, if the user has a question to communicate with the developer, you can move the mouse to a developer's avatar to get its mailbox.
 (3) News: This section of users can view our development log, to view all please click more. In all logs, users can click on the corresponding logs for different years.
(3) News: This section of users can view our development log, to view all please click more. In all logs, users can click on the corresponding logs for different years.

 (4) Visitors: The RevolverMaps widget shows users where visitors are located.
(4) Visitors: The RevolverMaps widget shows users where visitors are located.
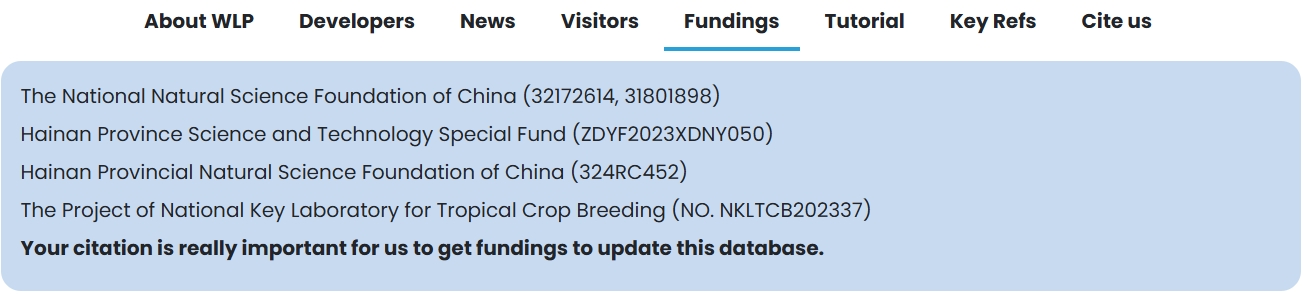
 (5) Fundings: This module provides the WLP project name and its serial number.
(5) Fundings: This module provides the WLP project name and its serial number.
 (6) Tutorial: This module summarizes the WLP tutorial and can be quickly moved to the tutorial page by clicking on Details.
(6) Tutorial: This module summarizes the WLP tutorial and can be quickly moved to the tutorial page by clicking on Details.
 (7) Key Refs: This module lists some of the top journals on water lilies, more journals can be viewed by clicking on Details or References in the Library in the navigation bar.
(7) Key Refs: This module lists some of the top journals on water lilies, more journals can be viewed by clicking on Details or References in the Library in the navigation bar.
 (8) Cite us: Location of Water Lily Pond.
(8) Cite us: Location of Water Lily Pond.

3. Hot tools!/Key horticultural genes!/Economic values
Click on different blocks to quickly jump to the more popular functions/different types of horticultural data query interface/different economic value modules of water lilies.
1. Nymphaeales
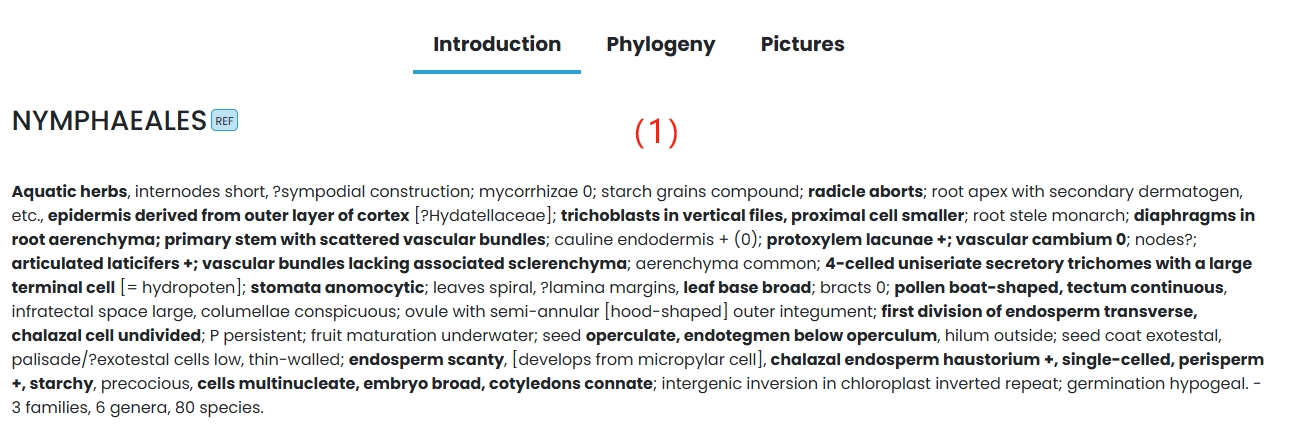
(1) Introduction: The whole nymphaea order is introduced in detail.
(2) Phylogeny: Tree diagram of basic information of nymphaeae composition.
(3) Pictures: A quick link map of several species assembled in WLP.


2. Hydatellaceae
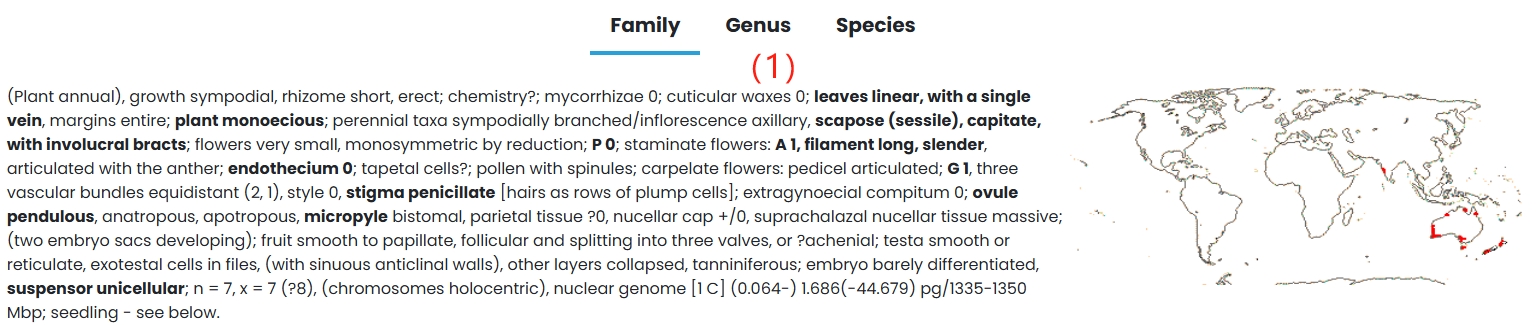
(1) Family: Hydatellaceae family is introduced in detail.
(2) Genus: A detailed introduction to the genus Trithuria.
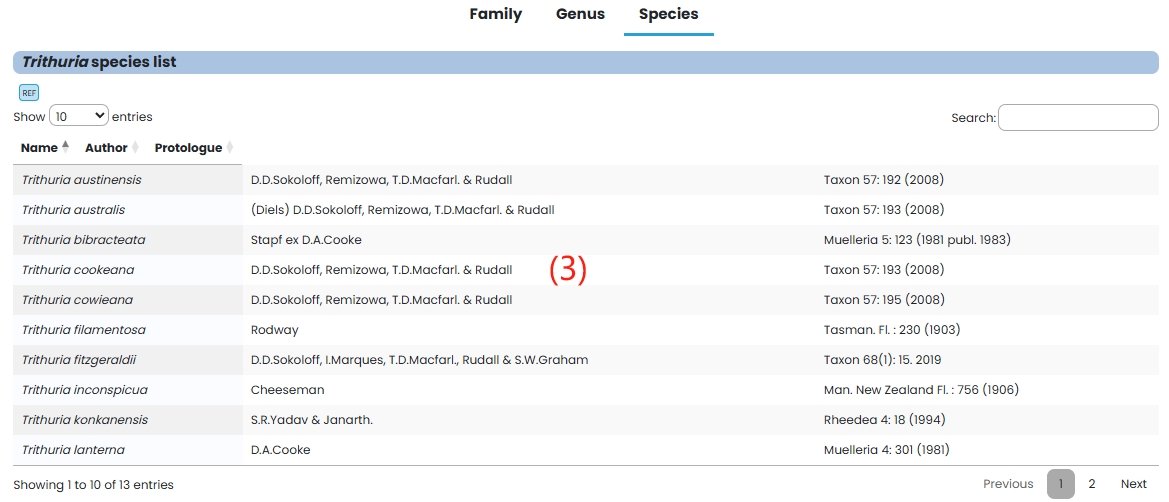
(3) Species: Species in the Hydatellaceae family are listed in tabular form.

3 Cabombaceae
(1) Family: Cabombaceae family is introduced in detail.
(2) Genus: A detailed introduction to the genus Cabomba and Brasenia.
(3) Species: Species in the Hydatellaceae family are listed in tabular form.
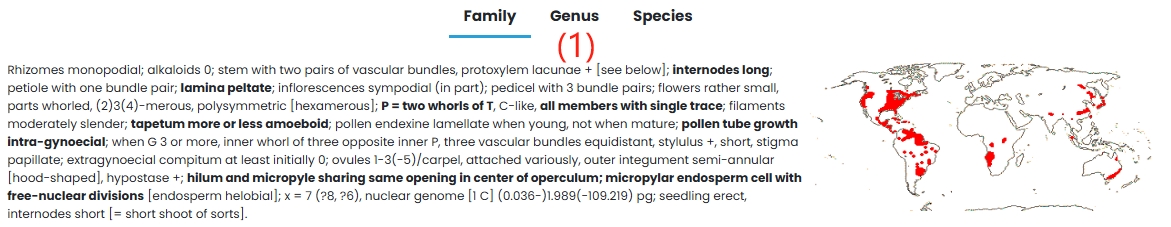
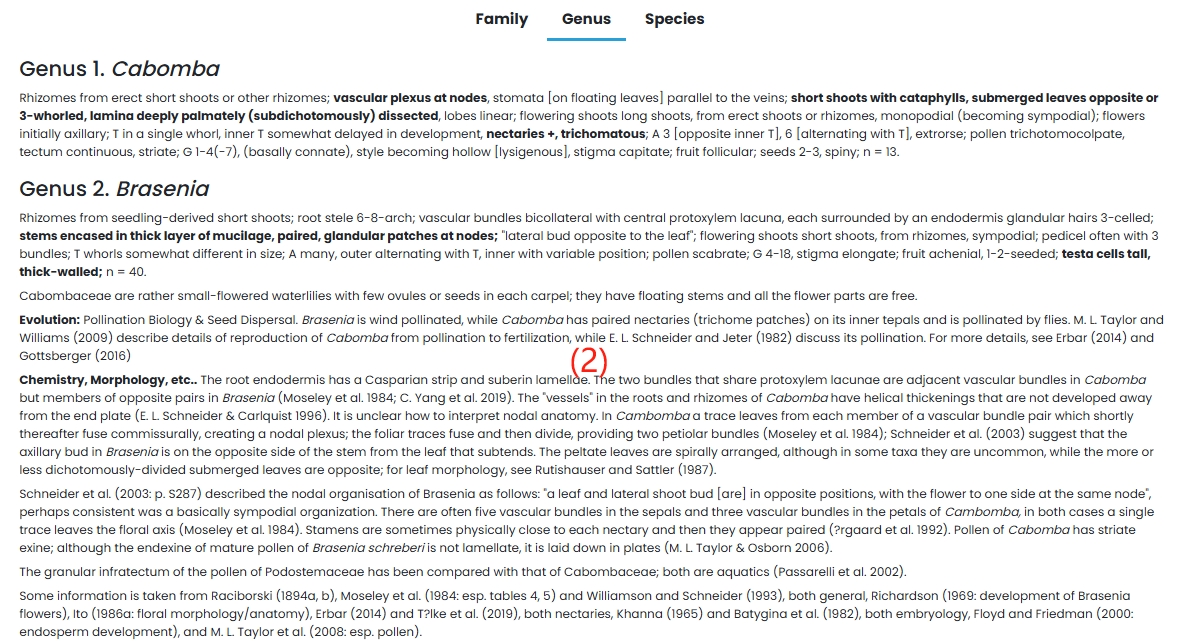
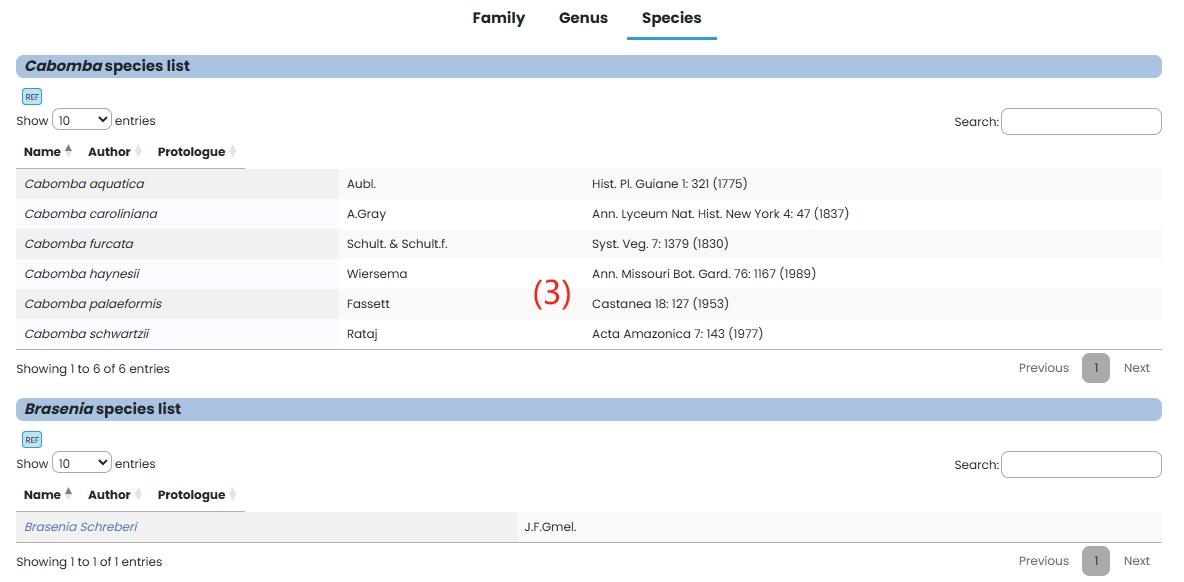
4. Nymphaeaceae
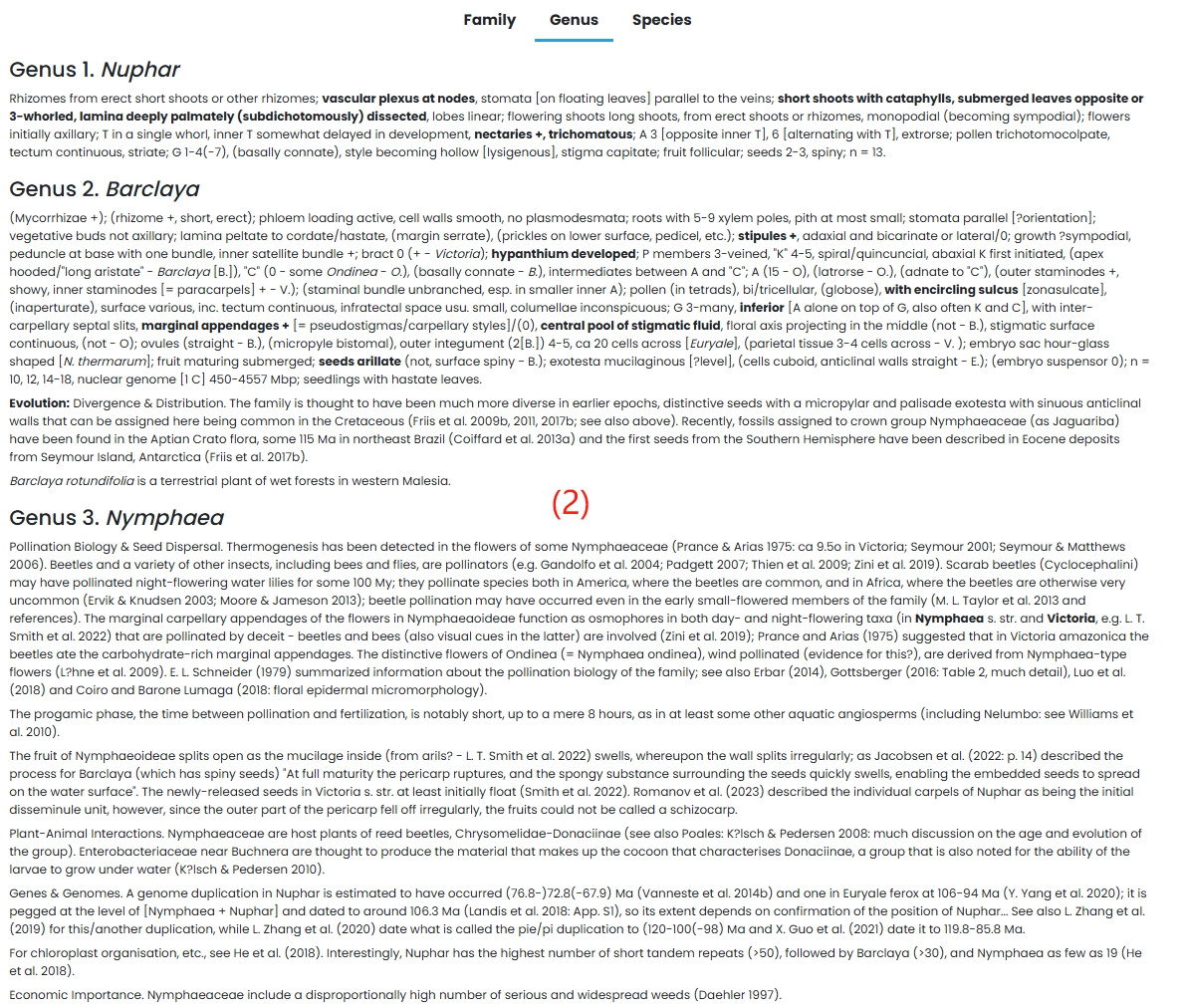
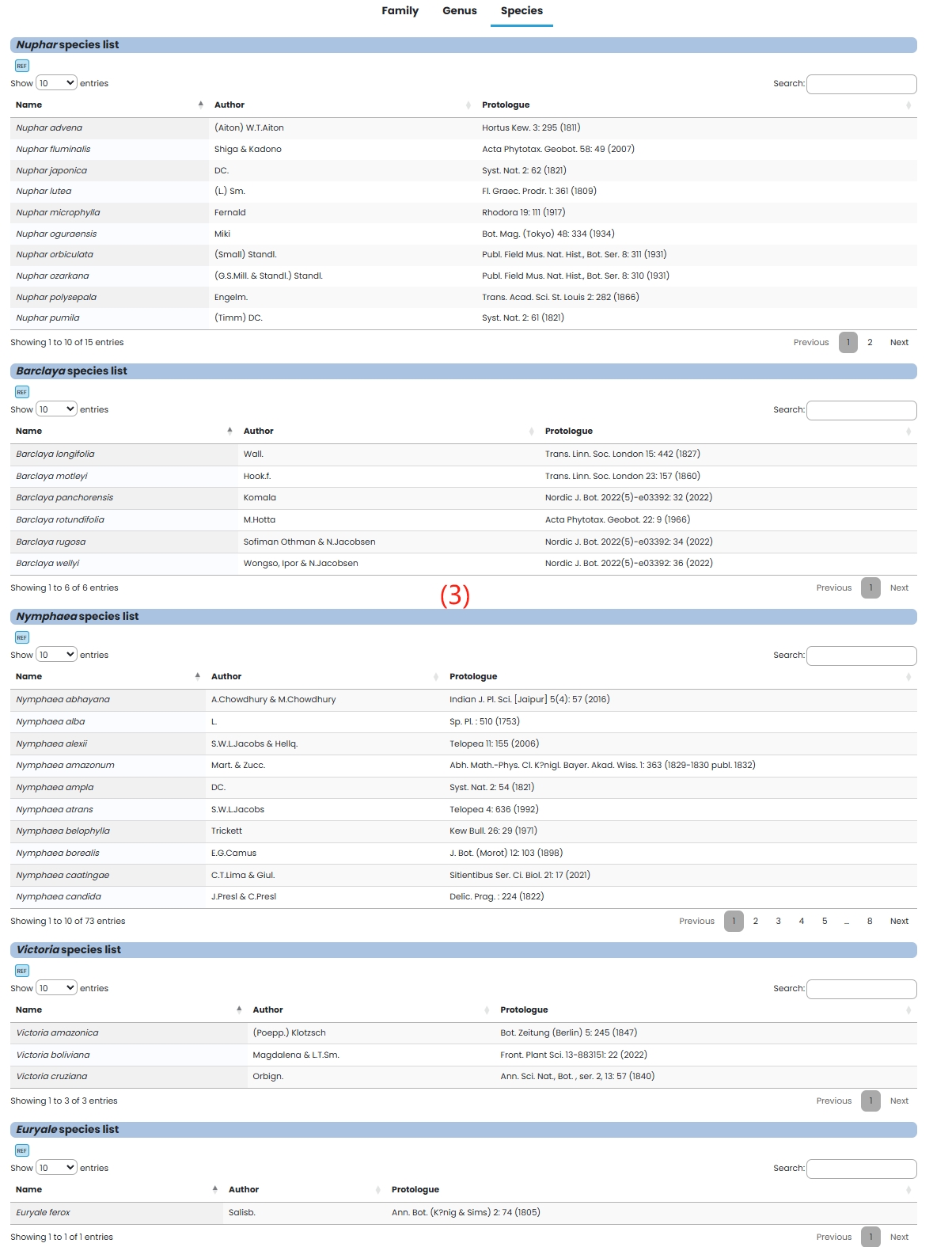
(1) Family: Nymphaeaceae family is introduced in detail.
(2) Genus: A detailed introduction to the genus Nuphar, Barclaya and Nymphaea.
(3) Species: Species in the Nymphaeaceae family are listed in tabular form.
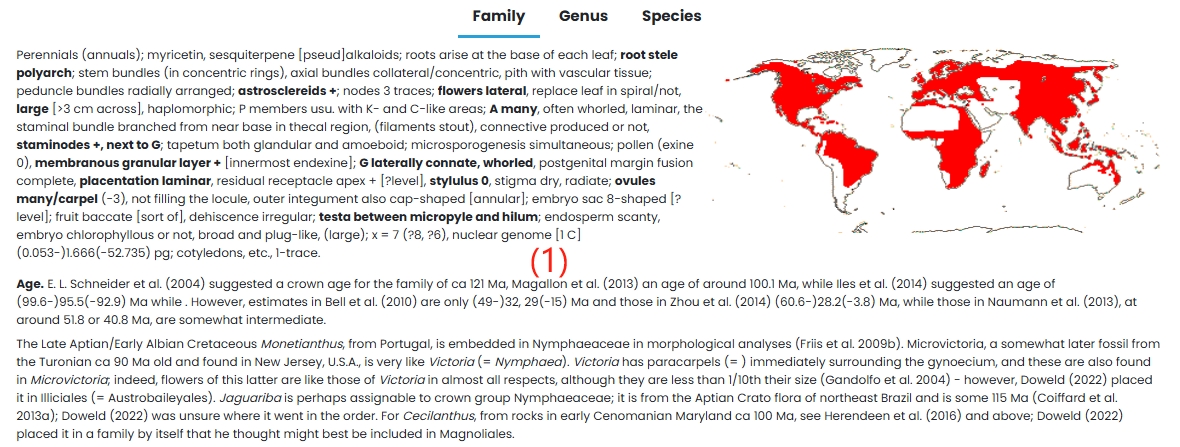
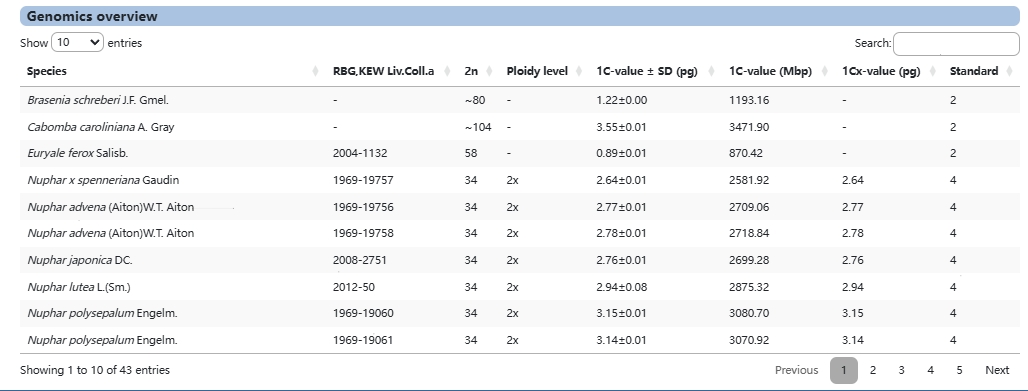
1. Overview
Overview presents users with genomic information for 48 different water lily species.
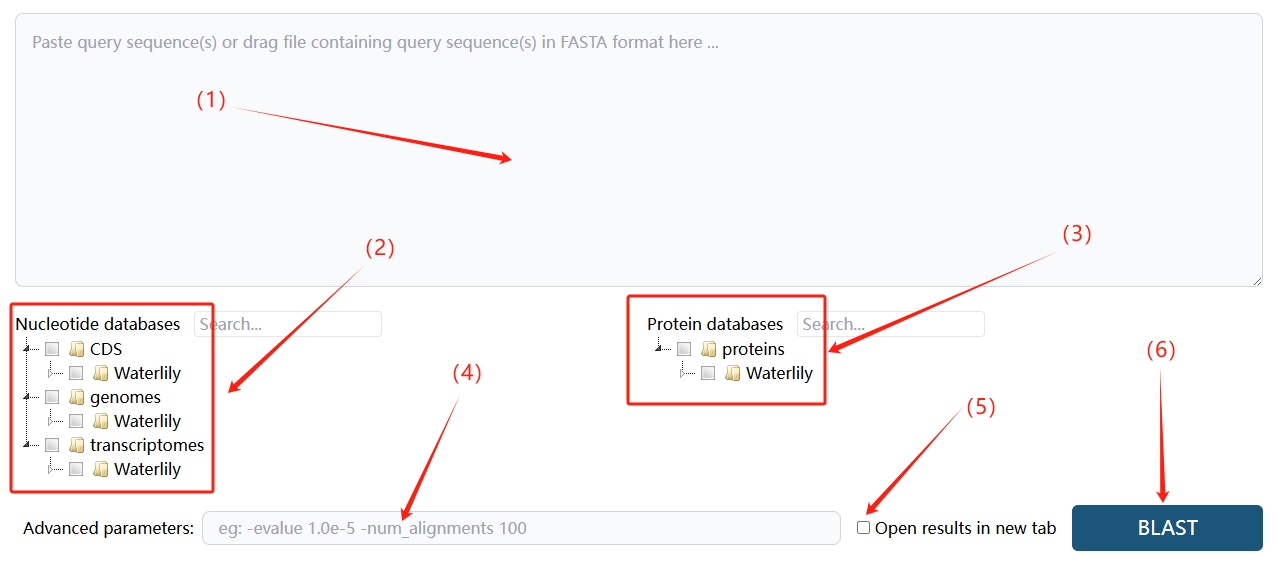
2. BLAST
(1) First, paste your query sequence(s) into the large text box at the center of the page or drag and drop a file containing sequences in FASTA format.
(2) On the left side of the page, under the "Nucleotide databases" section, select the desired database, such as "CDS," "genomes," or "transcriptomes" within the "Waterlily" directory.
(3) On the right side of the page, in the "Protein databases" section, choose the appropriate protein database, like the "Waterlily" directory under "proteins."
(4) If needed, customize your search using the "Advanced parameters" input box. Here, you can specify detailed options such as:
I. E-value threshold: This sets the maximum acceptable e-value for reported alignments, for example, -evalue 1e-5 to filter only significant matches.
II. Number of alignments: Control how many top alignments are displayed, for instance, -num_alignments 100 to show the top 100 matches.
III. Word size: Adjust the size of the initial matching word for a more fine-tuned search, e.g., -word_size 11.
IV. Gap penalties: For more customized gap scoring, you can use parameters like -gapopen 5 -gapextend 2.
(5) To view the results in a new tab, check the "Open results in new tab" option.
(6) Once all settings are configured, click the "BLAST" button at the bottom right to start the sequence alignment and obtain your results.
3. JBrowse
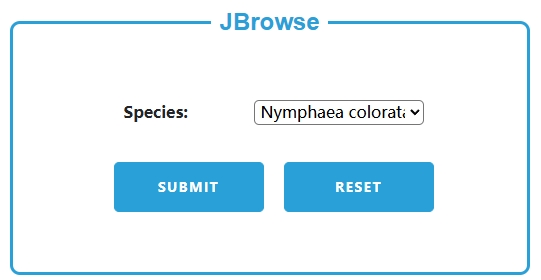
 (1) Select a species (e.g., Nymphaea colorata), and click submit to enter the corresponding JBrowse interface.
(2) In the JBrowse interface:
(1) Select a species (e.g., Nymphaea colorata), and click submit to enter the corresponding JBrowse interface.
(2) In the JBrowse interface:
I. Empty: This option allows users to start a new session without any preloaded data or settings. Users can add their own datasets and configuration files to create a fully customized genome browsing environment.
II. Linear Genome View: This option is used for viewing and browsing genome sequences and their annotations in a linear format. Users can explore different genes, variant sites, coverage data, and more, making it a core tool for detailed genome exploration.
III. Structural Variant Inspector: This tool is used for inspecting and visualizing structural variants in the genome, such as chromosomal rearrangements, insertions, and deletions. It provides users with a deeper analysis perspective to help identify and study complex structural variations in the genome.
4. Gene search
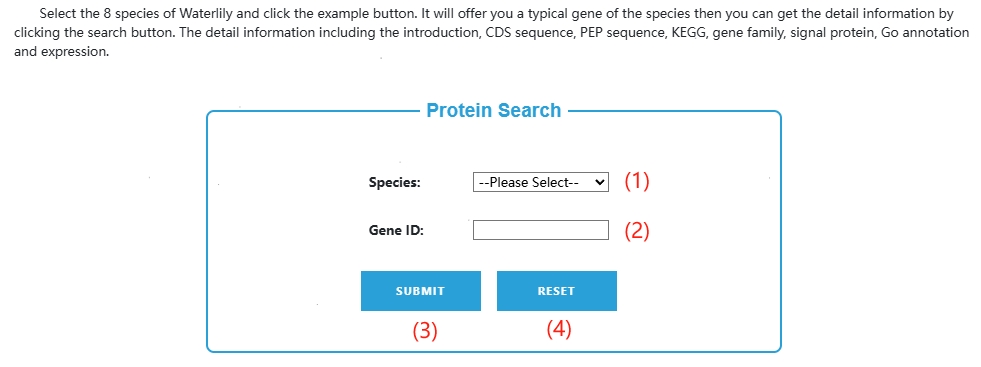
 (1) Click here to select the species you want to query.
(2) After selecting the species in (1), the sample id of the corresponding species will be automatically filled in here, or manually delete the sample and fill in the id you want to query.
(3) After filling in (1) (2), click this button to jump to the result page.
(4) Click this button to quickly clear everything filled.
(1) Click here to select the species you want to query.
(2) After selecting the species in (1), the sample id of the corresponding species will be automatically filled in here, or manually delete the sample and fill in the id you want to query.
(3) After filling in (1) (2), click this button to jump to the result page.
(4) Click this button to quickly clear everything filled.
5. Synteny search
The Synteny offers pairwise collinear comparisons across six species, allowing users to select species and chromosomes for collinearity mapping.
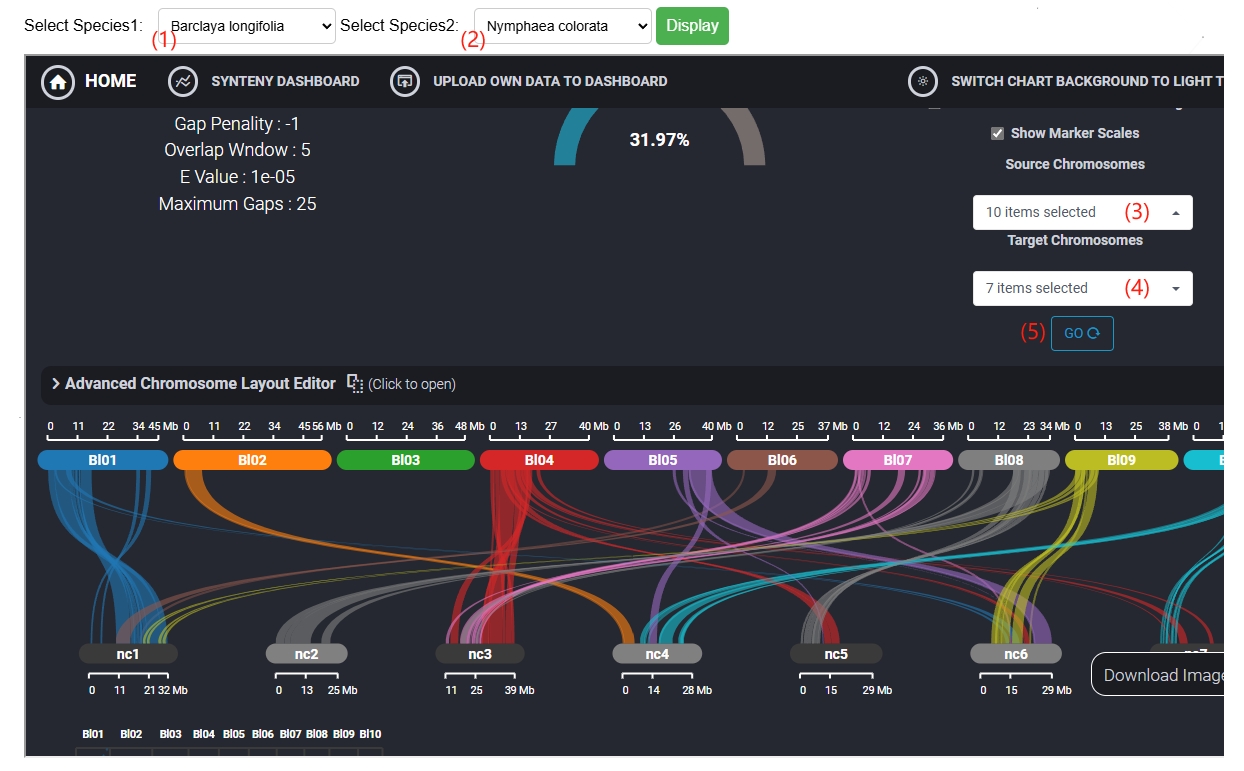 (1) Choose the first species here.
(2) Choose the second species here.
(3) Select the chromosomes of the first species.
(4) Select the chromosomes of the second species.
(5) Clicking here will bring up synteny results.
(1) Choose the first species here.
(2) Choose the second species here.
(3) Select the chromosomes of the first species.
(4) Select the chromosomes of the second species.
(5) Clicking here will bring up synteny results.
6. Gene Family Search
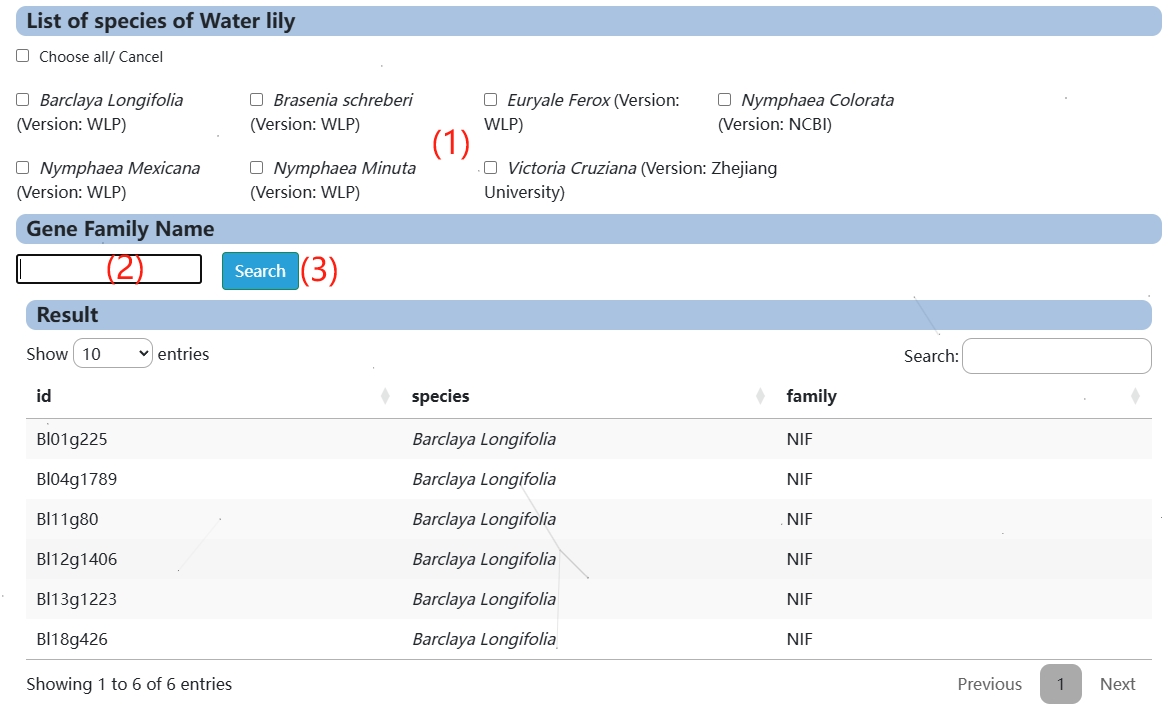 (1) Select the species you want to query, either by selecting one or more species, or by clicking Choose all/ Cancel to select all or deselect all.
(2) Click here to automatically fill in the sample family name or the family name you want to query.
(3) Clicking Search will display the query results.
(1) Select the species you want to query, either by selecting one or more species, or by clicking Choose all/ Cancel to select all or deselect all.
(2) Click here to automatically fill in the sample family name or the family name you want to query.
(3) Clicking Search will display the query results.
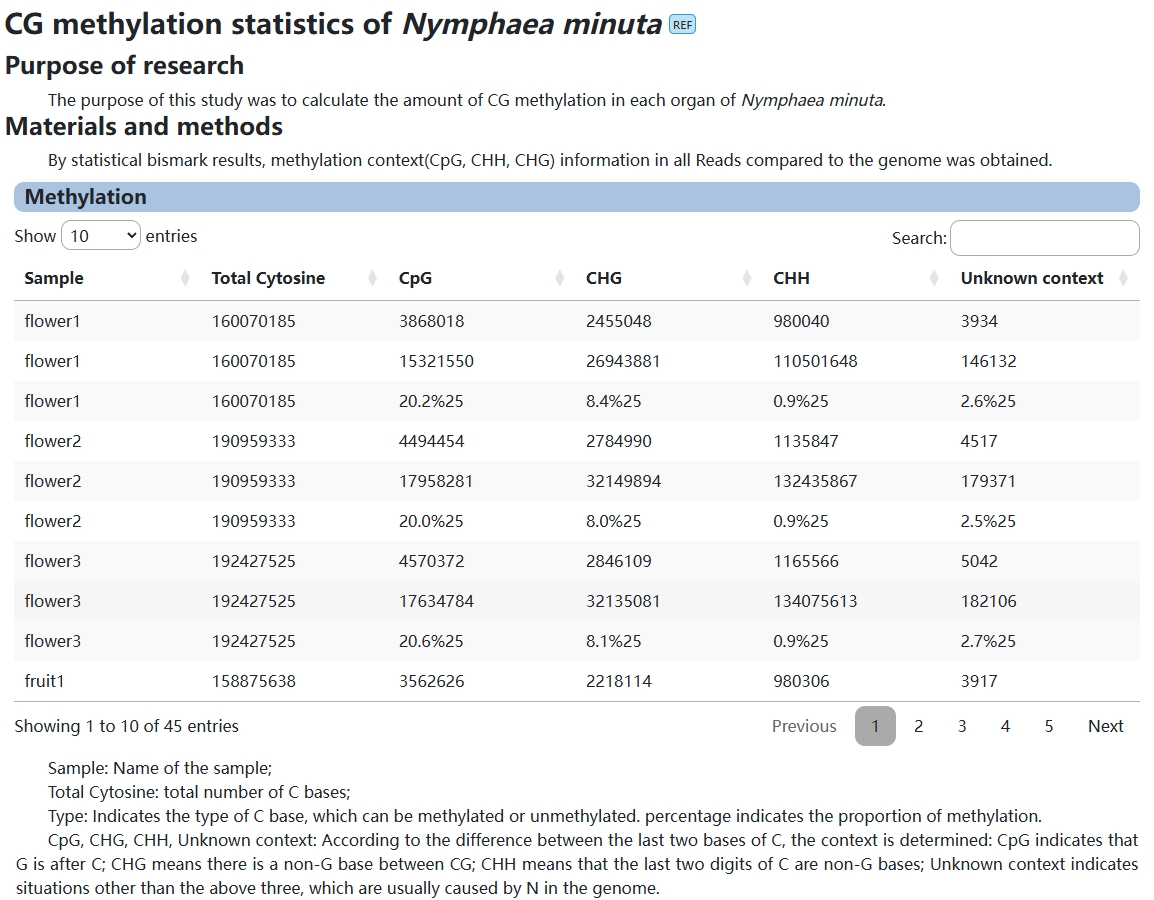
7. Methylation
This module shows the user information about methylation in Nymphaea minuta.
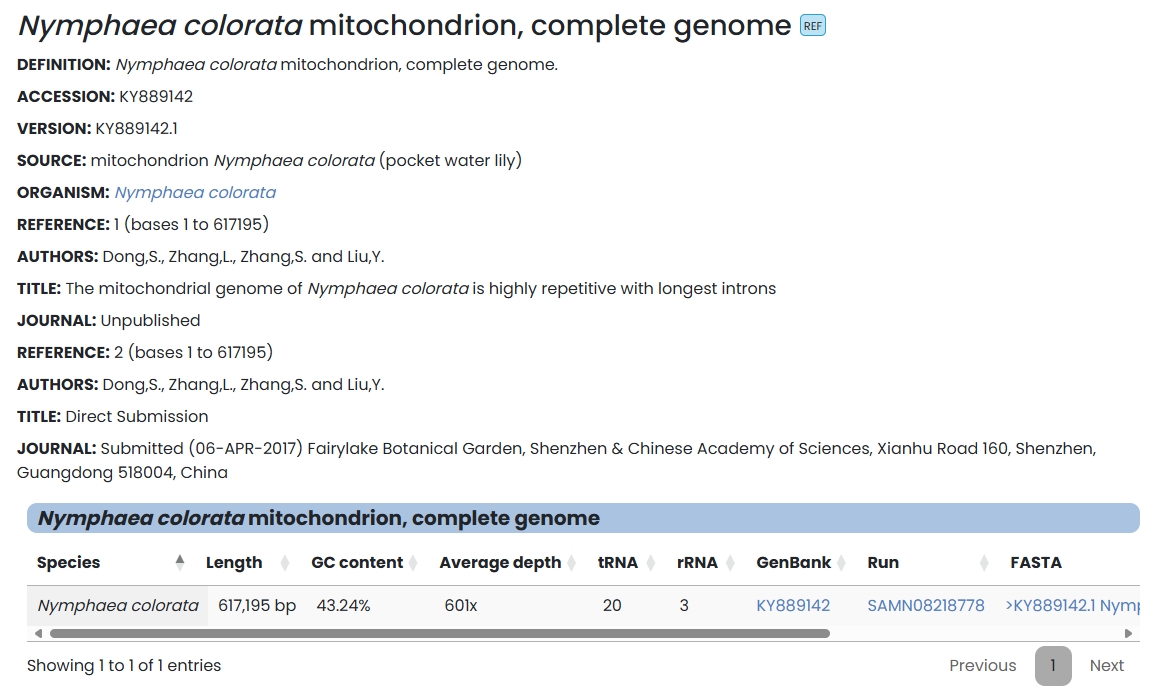
8. Mitochondrion
This module collects mitochondrial assembly information from multiple species. Users can click on the blue font in each species list to link directly to NCBI related content about the species' mitochondria, including: GenBank, Run, FASTA file, etc.
9. Chloroplast
This module collected chloroplast assembly information for multiple species. Users can click on the blue font in each species list to link directly to NCBI's chloroplast content for that species, including: GenBank, FASTA file, etc.

1. eFP-gene
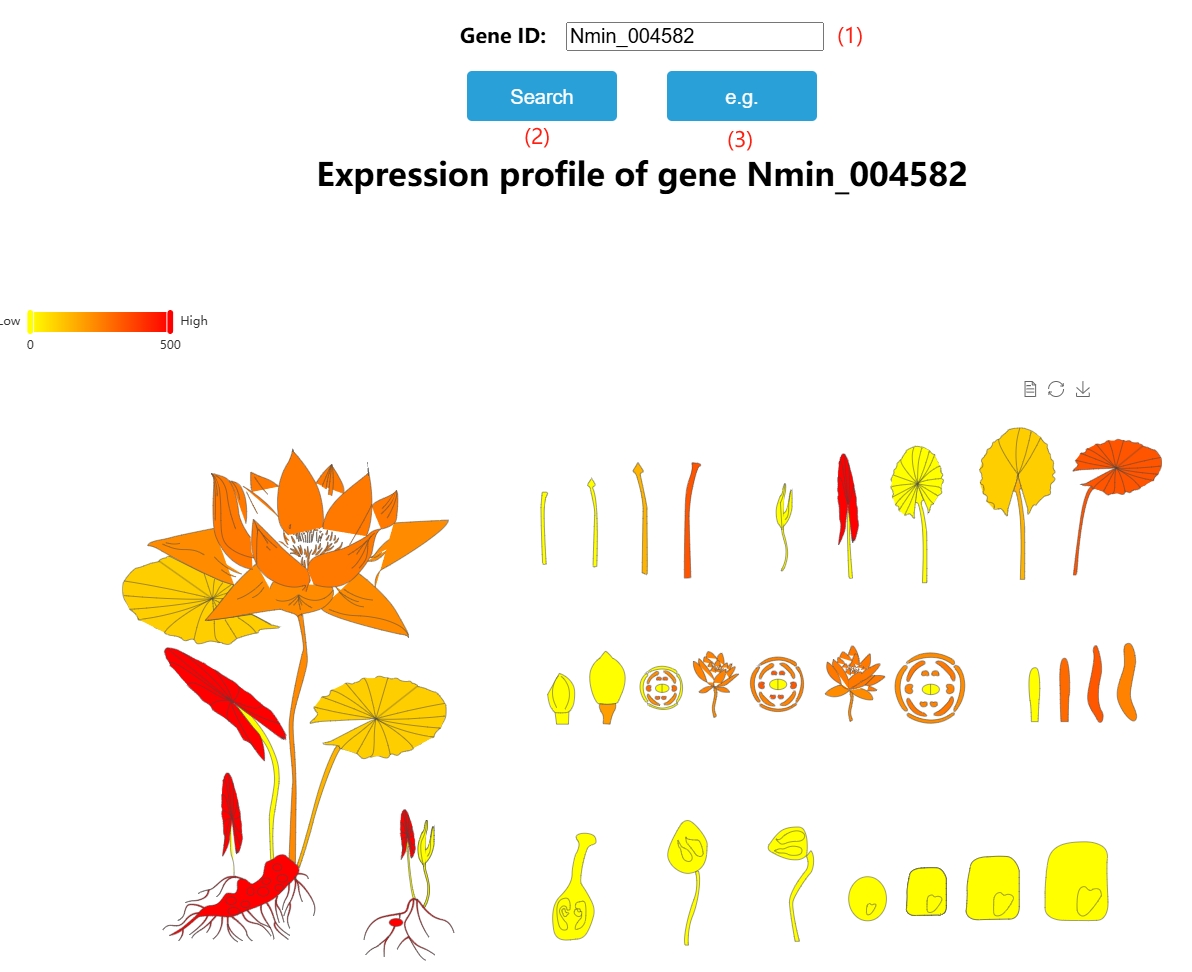 (1) Enter the id here.
(2) Click here for results.
(3) Click here to automatically fill in the sample id.
(1) Enter the id here.
(2) Click here for results.
(3) Click here to automatically fill in the sample id.
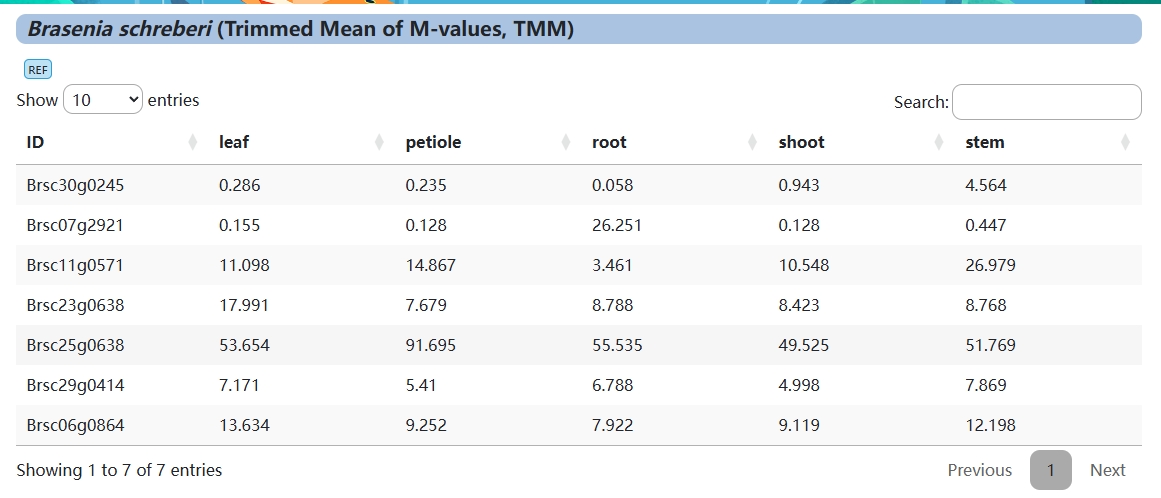
2. Expression value
Trimmed Mean of M-values, TMM
TMM is a method of standardization between samples.
TMM assumes that the expression of most genes does not vary between samples.
TMM normalizes total Reads Counts between samples and does not normalize gene length or library size
TMM considers sample RNA population and effective in normalization of samples with diverse RNA repertoires (e.g. TMM has certain advantages in processing batch effects of data when comparing samples from different tissues or genotypes, or when there are significant differences in RNA Population between samples.
TMM performs better in comparison between samples and can be implemented using R package: edgeR.
edgeR assumed that gene length was constant from sample to sample and did not consider normalizing gene length.

 (1) Select species here.
(2) After selecting a species in (1), the id set is automatically filled here. You can manually delete the example and fill in the id you want to query.
(3) Click this button to jump to the query result.
(4) Click this button to clear the selected and filled information.
(1) Select species here.
(2) After selecting a species in (1), the id set is automatically filled here. You can manually delete the example and fill in the id you want to query.
(3) Click this button to jump to the query result.
(4) Click this button to clear the selected and filled information.
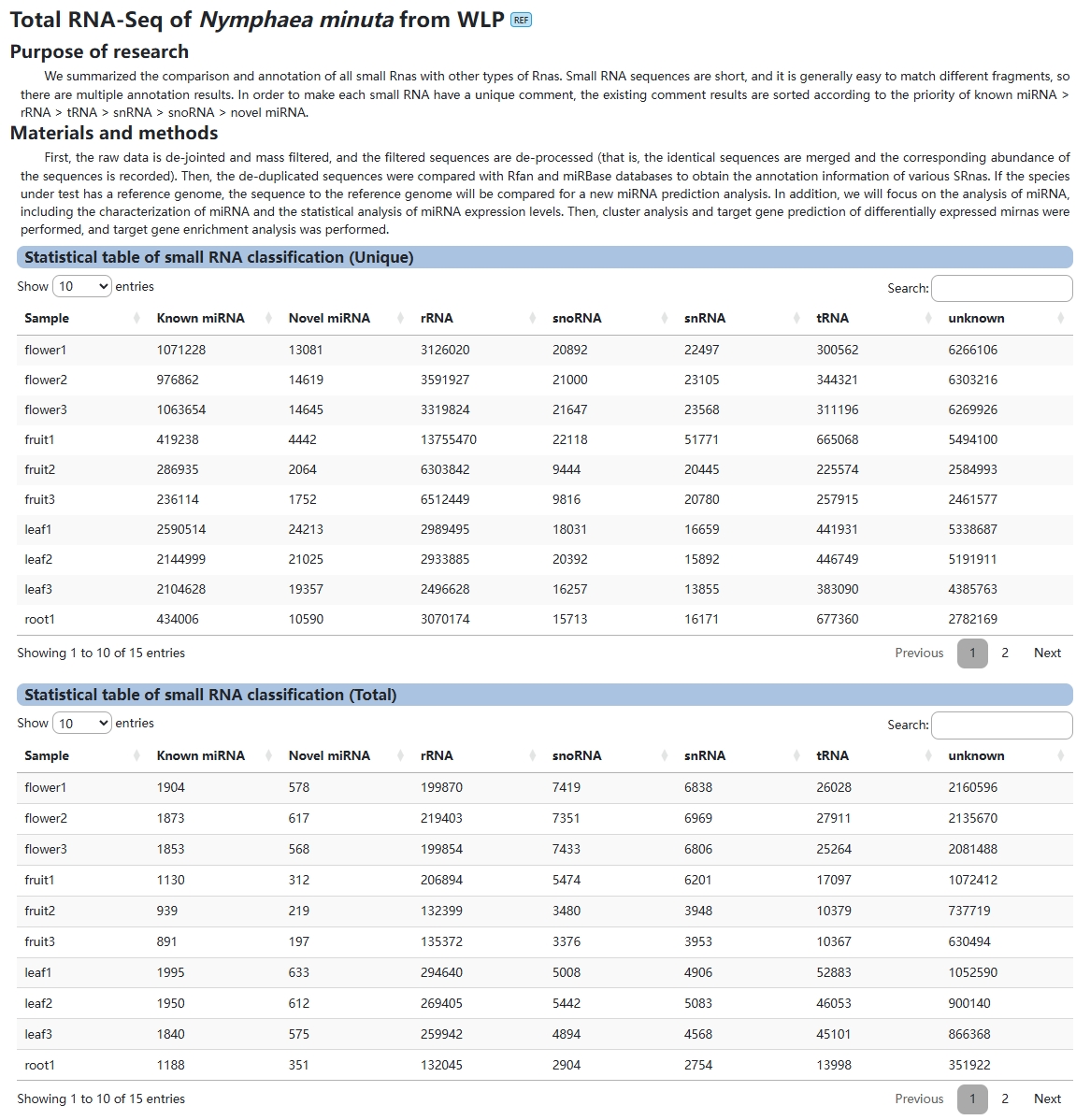
3. N. minuta Total RNA-Seq
This module provides the user with full transcriptome information on Nymphaea minuta.
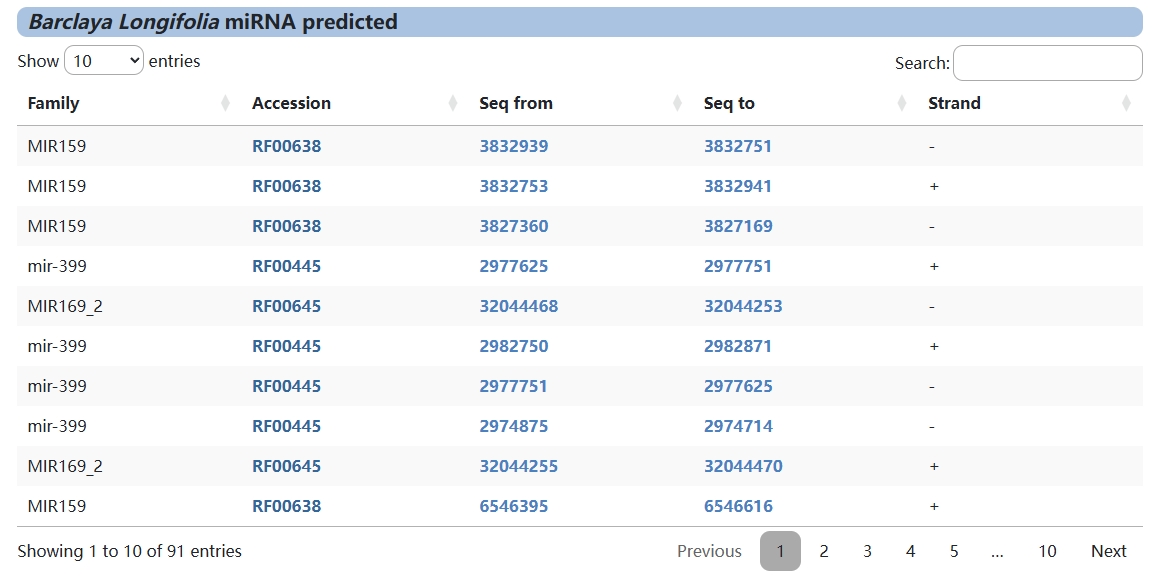
4. miRNA
 (1) Select the species you want to query.
(2) Click here to go to the query results page for the species selected in (1).
(3) Click here to clear the selection.
(1) Select the species you want to query.
(2) Click here to go to the query results page for the species selected in (1).
(3) Click here to clear the selection.
5. lncRNA
The purpose of this module is to provide users with information about lncrnas in Nymphaea minuta.
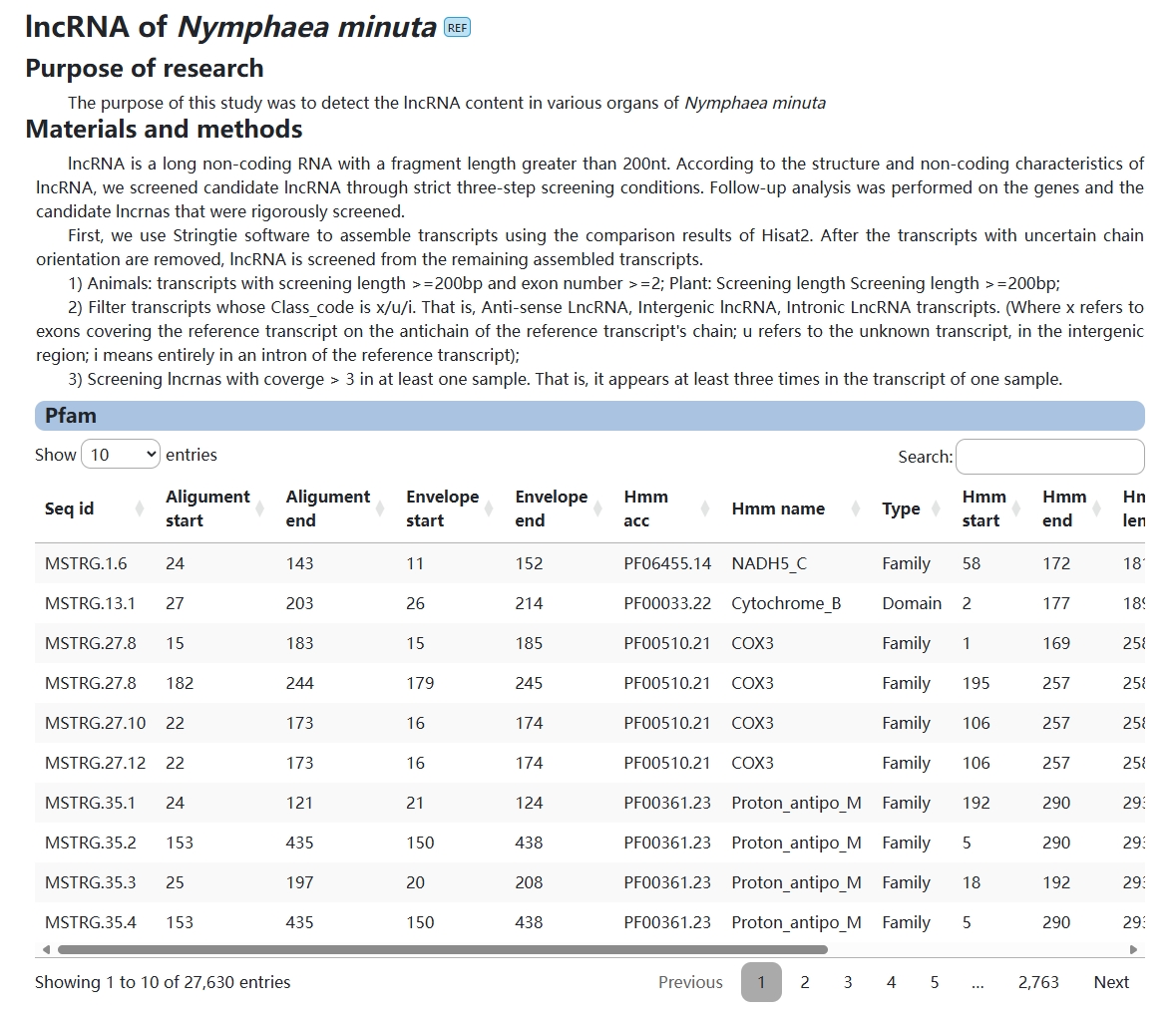
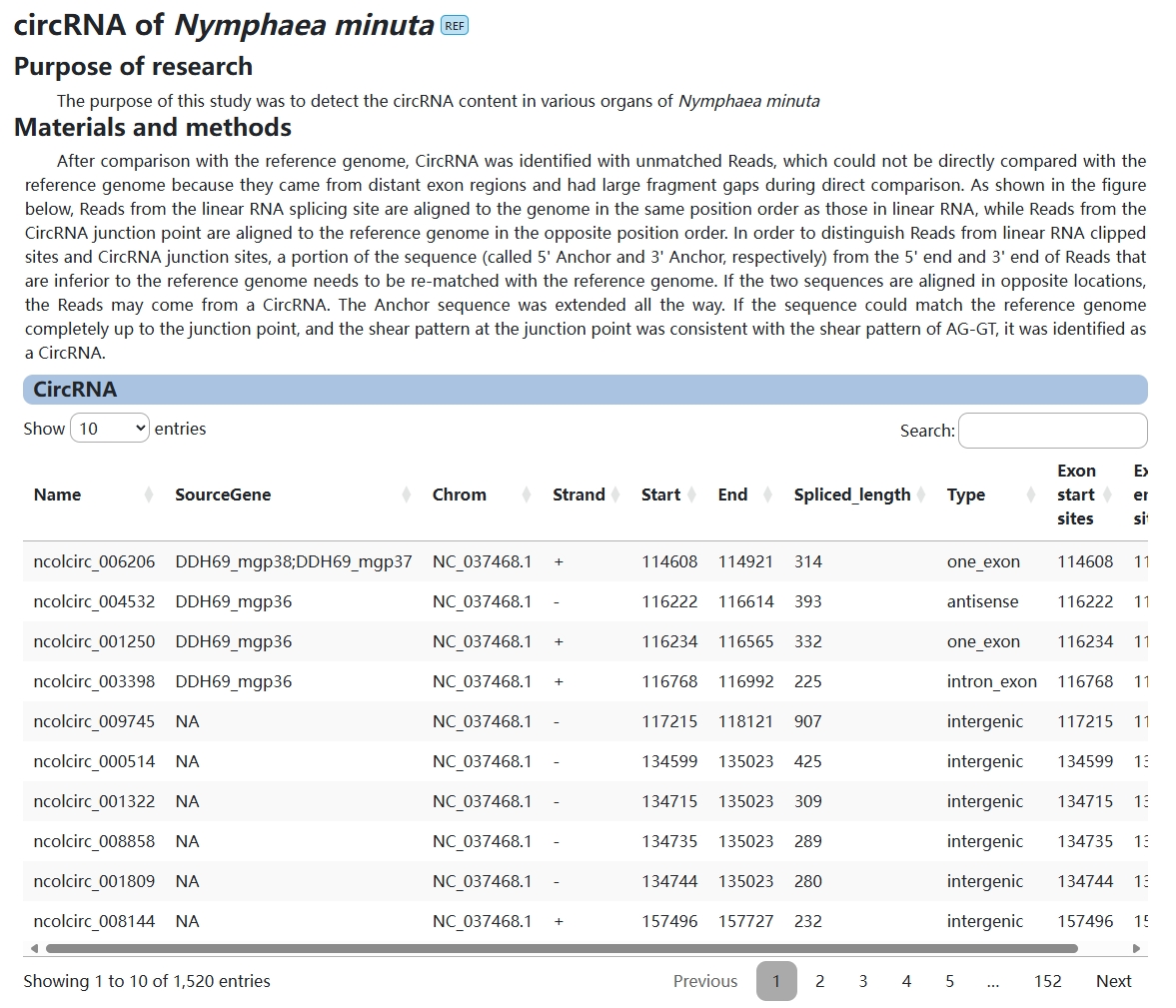
6. circRNA
The purpose of this module is to provide users with information about circRNA in Nymphaea minuta.
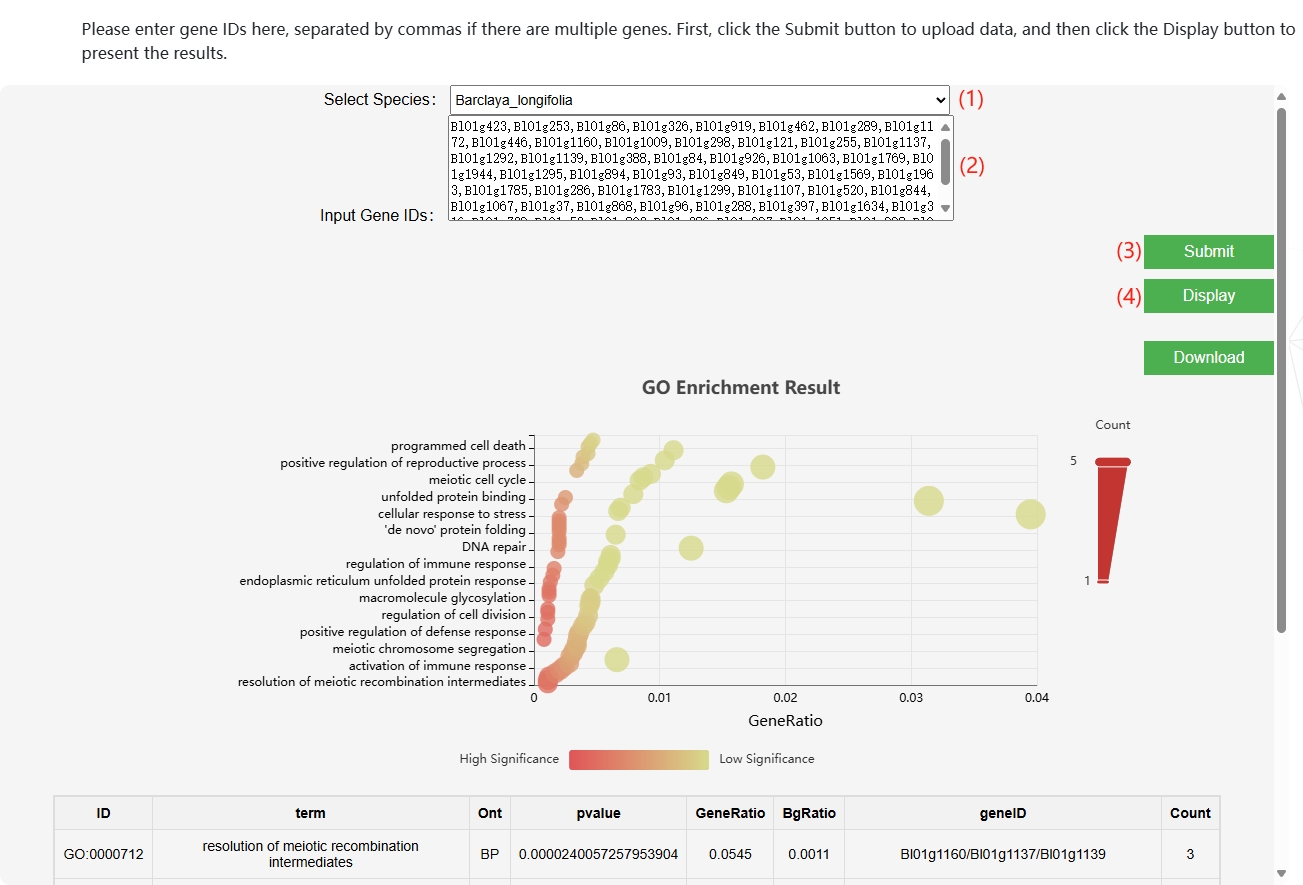
1. GO Enrichment/ KEGG Enrichment
This module integrates the GO databases of eight species, allowing users to input gene IDs for GO enrichment analysis and generate GO enrichment bubble plots.
 This module incorporates the KEGG databases of six species, enabling users to input gene IDs for KEGG enrichment analysis and generate KEGG enrichment bubble plots.
This module incorporates the KEGG databases of six species, enabling users to input gene IDs for KEGG enrichment analysis and generate KEGG enrichment bubble plots.
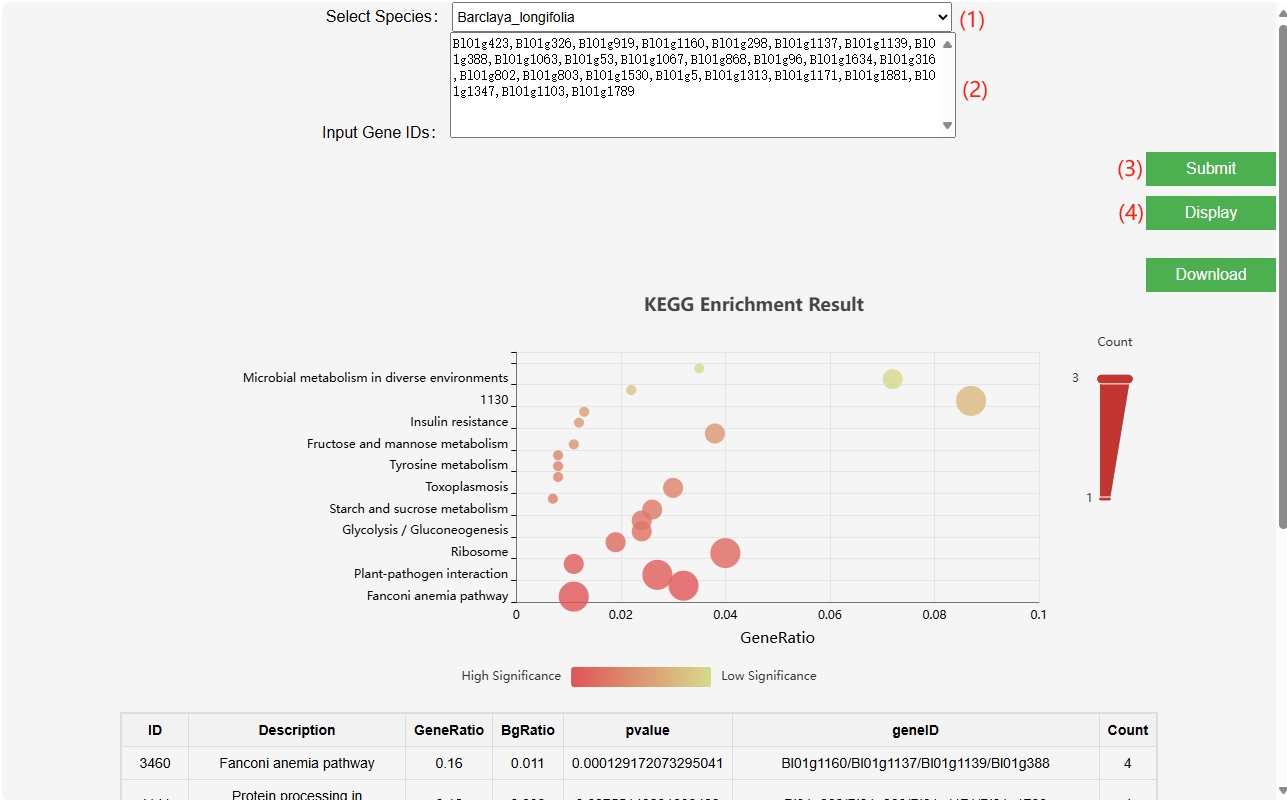 (1) Select the species you want to analyze.
(2) After selecting a species in (1), the sample id will be automatically filled in this field, or you can delete the sample id and enter the id you want to analyze.
(3) When (1) (2) is completed, click this button to confirm submission.
(4) Click (3) to confirm submission and then click this button to display the analysis results.
(1) Select the species you want to analyze.
(2) After selecting a species in (1), the sample id will be automatically filled in this field, or you can delete the sample id and enter the id you want to analyze.
(3) When (1) (2) is completed, click this button to confirm submission.
(4) Click (3) to confirm submission and then click this button to display the analysis results.
2. Transcription Factor
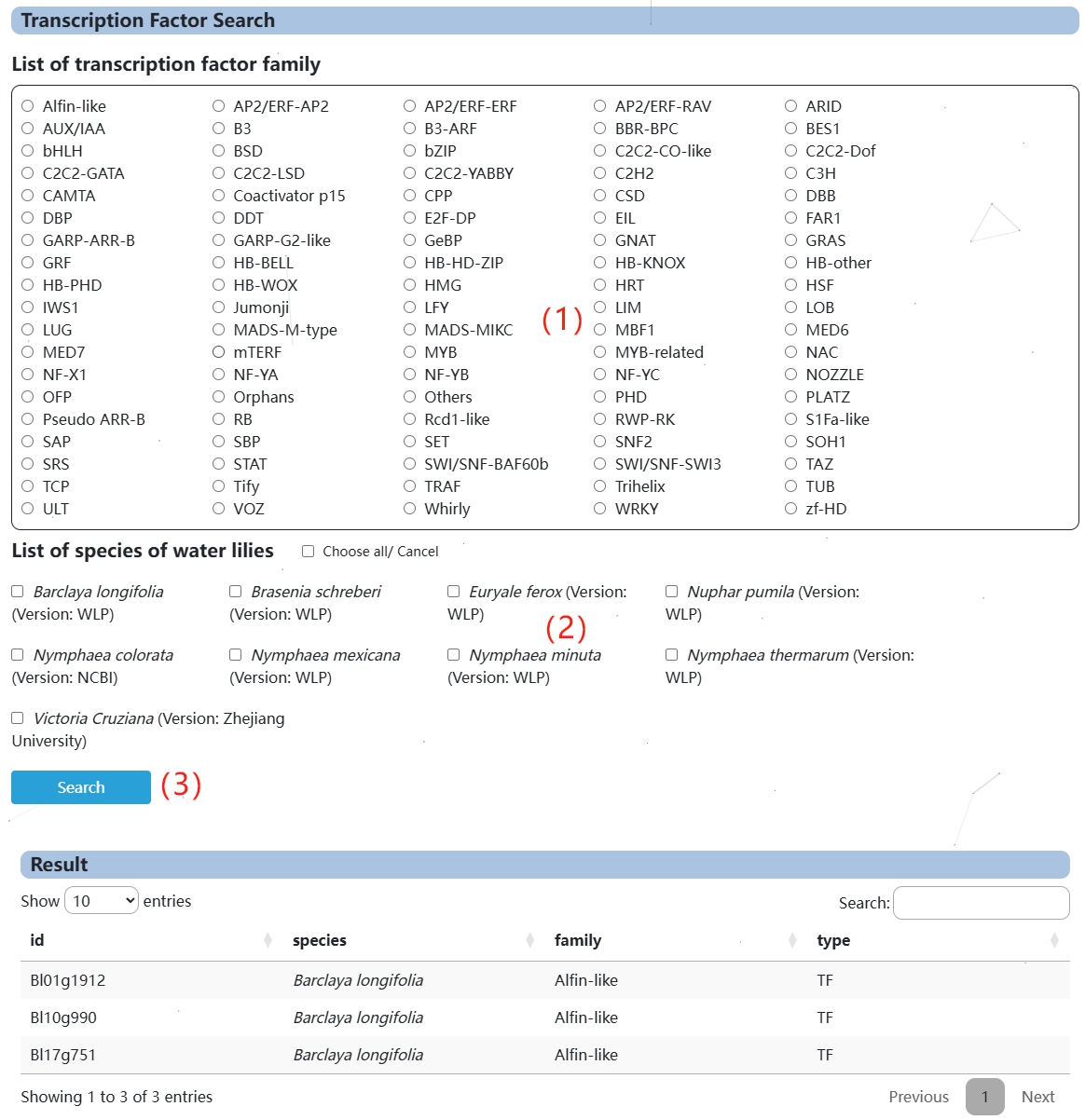 (1) In this section you can select the transcription factor you want to query.
(2) Select the species you want to query here, one or more.
(3) Clicking Search will display the query results.
(1) In this section you can select the transcription factor you want to query.
(2) Select the species you want to query here, one or more.
(3) Clicking Search will display the query results.
3. N. minuta proteomics
This module provides users with the proteome data and various analytical diagrams of Nymphaea minuta.

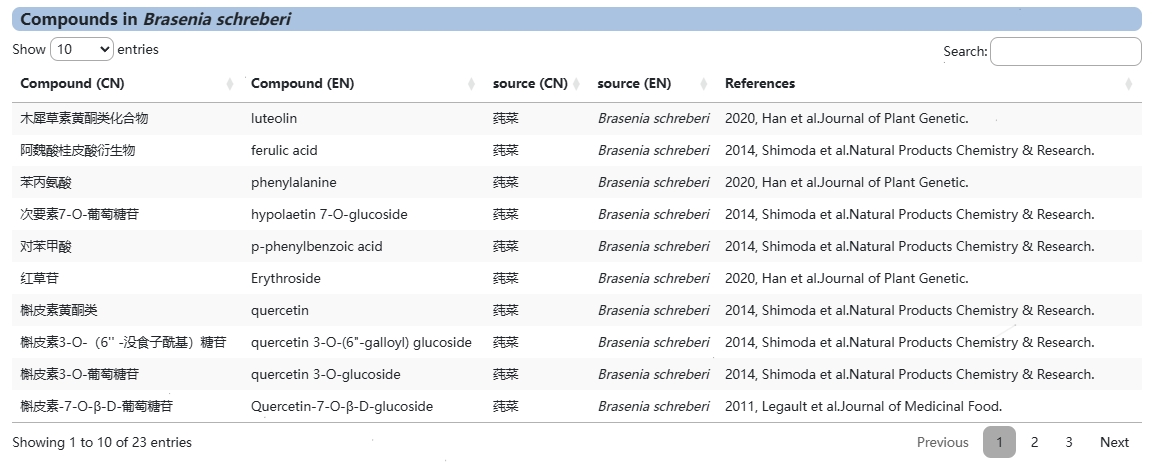
This module provides users with metabolite information for most species of Nymphaea, Brasenia, Nuphar and species Nymphaea minuta, using Brasenia as an example.
1. Cultivars
This module presents the user with phenotypic information on the flowers and leaves of 180 cultivated species.

2. Species
The module shows users phenotypic pictures of some of the original species' root leaf flowers and other organs.

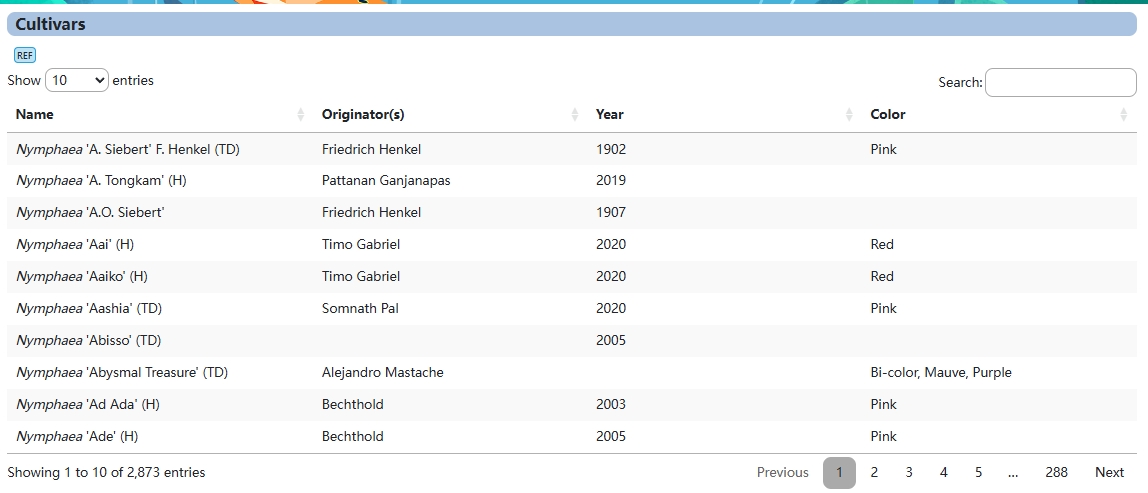
1. Cultivars Breeding
This module provides users with variety information on more than 2,800 nymphaeae species.
2. Molecular. Breeding
This module provides users with some information about water lily molecular breeding.

3. Products
This section shows users the products related to water lilies.
4. Cultivation and management
This module shows users some of the main pest and disease pictures about water lilies.
1. References
This module provides the user with the available published literature on water lilies.
2. References
The module provides users with some patents on water lilies.
3. Bookss
The module provides users with some books on water lilies.
4. Data
The module provides users with some downloadable water lily data.