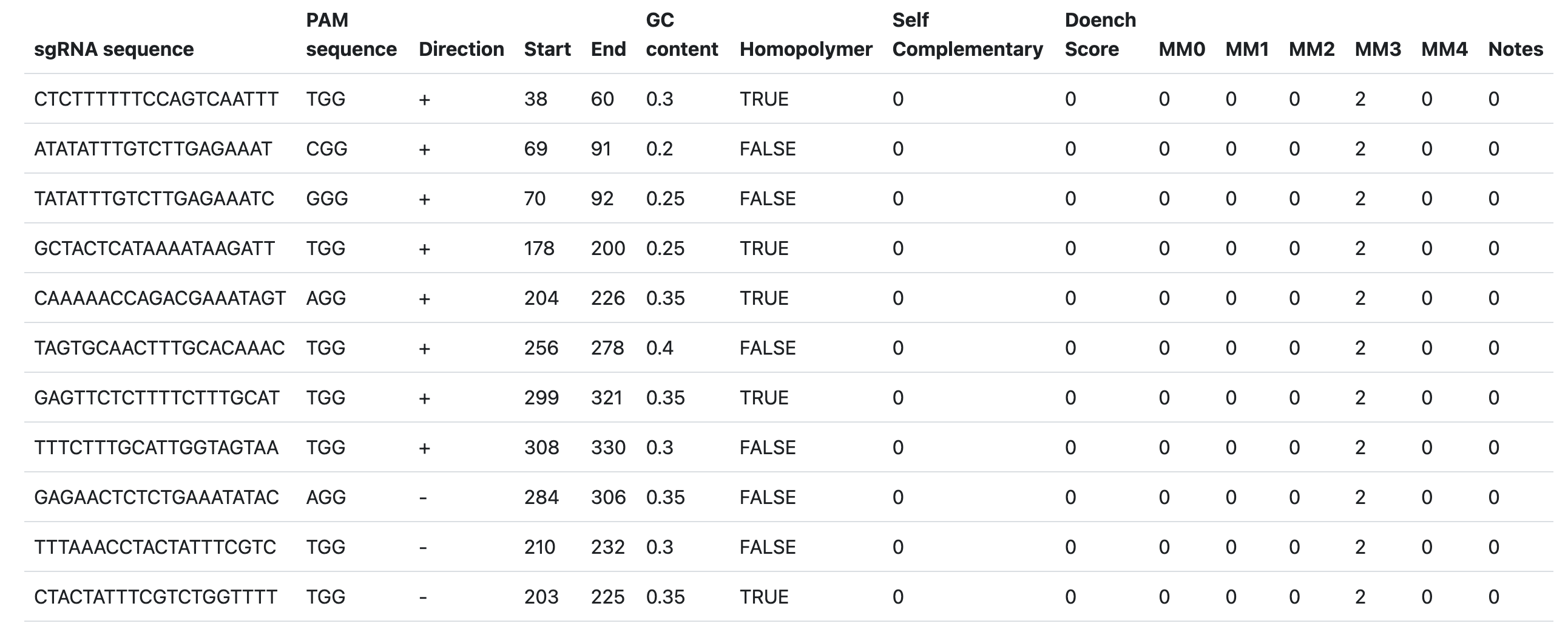
GuideRNA
A tool for designing optimal guide RNAs (gRNAs) for CRISPR-Cas genome editing experiments. The tool identifies potential target sites within a genome and evaluates them to minimize off-target effects, which is crucial for precise gene editing. Its applications include analyzing genomics data, gene function annotation data, and transcription factor binding site data to achieve accurate gene knockouts or modifications.
Input:
.gtf, .csv, .xls, .xlsx, .xlsm, .txt
Sequence:
ATCTTCTCATGCTTCTGTATGTGATTGTTTTTCTTCGCTCTTTTTCCAGTCAATTTTGGTGCAATGCATATATTTGTTCTTGAGAAATCGGGACTCCTTAAATCAATAGCCTTCTGTCAAAAGAGAGCTATCTTTCACCTTAGTGCTATGCCTAGCAAGCACTAGAGCTCCAATCATGCTACTCATAAAATAAGATTTGGAACCAAAAACCAGACGAAATAGTAGGTTTAAAGTAAATTGCTCAACGTATTCAAATTAGTAGCAACTTTGCACAAACTGGCATAACCGTATATTTCAGAGAGTTCTCTTTTCTTTGCATTGGTAGTAATGGAATGCTTTCATTGTCTAAAATGAAGAATATTTTACTATTTTGTAGTTGCACTTTTATCTATCATCTAAAAGTAACAATTCTGCTGCTTGATCATTTT
Output: